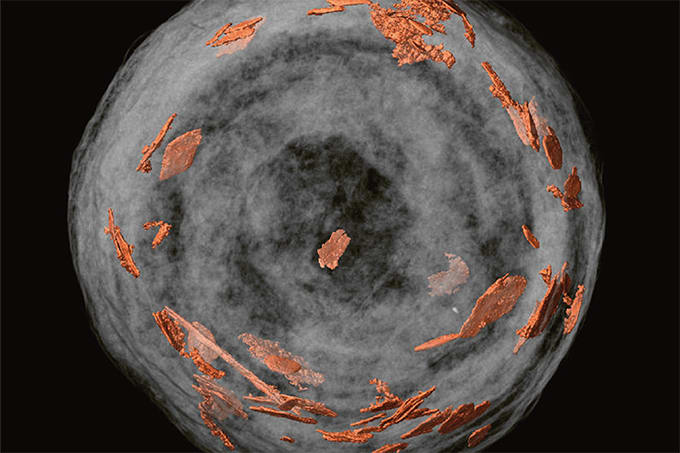
A new publicly accessible reference library, developed at the University of California San Diego, can detect drug exposure directly from patient samples – offering a new way to uncover medications and contaminants that might otherwise go unnoticed. Described in Nature Communications, the Global Natural Product Social Molecular Networking (GNPS) Drug Library combines high-resolution mass spectrometry with curated annotations to detect over 2,000 drugs, metabolites, and related compounds across complex biological matrices.
The tool addresses a long-standing gap in clinical data: patients often take over-the-counter drugs, leftover prescriptions, or supplements that go unrecorded in medical records. These omissions can hinder both treatment and research. By comparing molecular fragmentation patterns from biological samples to those in the GNPS Drug Library, researchers can directly identify drugs and metabolites that may be absent from medical records.
The team used tandem mass spectrometry (MS/MS) to generate characteristic fragmentation patterns of each compound. The database links each entry with metadata such as drug class, source (prescription, over-the-counter, environmental), mechanism of action, and therapeutic uses. A web interface enables clinical researchers without a pharmacology background to annotate their datasets with minimal effort.
“Whatever sample we put into the mass spectrometer, be it urine, breast milk or even an environmental water sample, it will be able to detect all of the chemicals in the sample,” said co-first author Nina Zhao, a postdoctoral researcher in the lab of Pieter Dorrestein.
In a proof-of-concept analysis of nearly 2,000 samples from the American Gut Project, the team detected 75 distinct drugs – reflecting the most commonly prescribed classes in the United States, Europe, and Australia. U.S. participants carried more detectable drugs per individual than their European or Australian counterparts. Further analysis revealed that painkillers were more often found in female participants, while erectile dysfunction drugs were primarily detected in males.
Additional case studies included patients with Alzheimer’s disease, HIV, and Kawasaki disease. For example, Alzheimer’s patients showed signatures of both cardiovascular and psychiatric medications. In HIV samples, the library revealed both antiviral and off-target drugs such as antidepressants, enabling researchers to cluster participants based on actual drug exposure rather than reported use.
By linking drug exposure to microbiome changes and uncovering hidden environmental contaminants, the GNPS Drug Library opens new avenues for personalized therapy and exposure monitoring.
“By understanding that, maybe we can use this information to optimize drug treatment,” Zhao added.