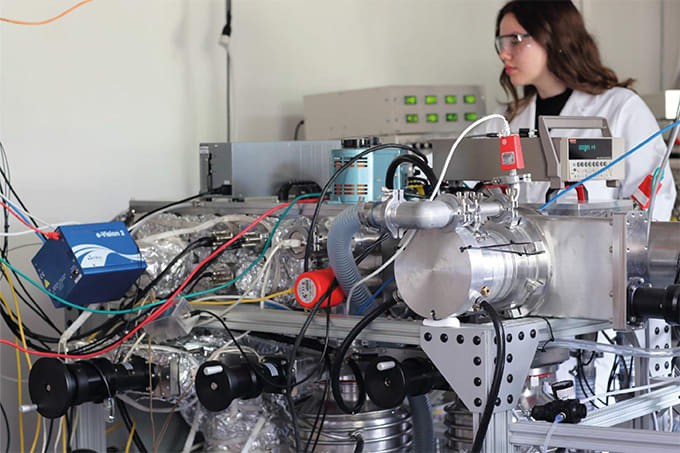
Digestion with the FabRICATOR® (IdeS) enzyme, which specifically cleaves IgG under the hinge region, followed by reduction results in 3 distinct mAb subunits (LC, Fd and Fc/2) in the range of 22-27 kDa. Optionally, selective Fc-deglycosylation can be performed using IgGZERO™ (EndoS) or GlycINATOR™ (EndoS2) to further reduce heterogeneity. Either of the two enzymes can be co-incubated with FabRICATOR ® during the digestion step. They facilitate characterization of posttranslational modifications like glycan heterogeneity, oxidation, lysine clipping and a rapid confirmation of amino acid sequence. The method can be applied for tasks like clone selection, process development, formulation and stability studies of monoclonal antibody based bio therapeutics such as MAb’s, Fc-fusion proteins, biosimilars and ADC’s.

Here we present a two-step process, which can be easily automated, for the preparation and analysis of antibody subunits by ESI-QTOF. By using this method for applications like sequence verification by accurate mass, sample preparation time can be reduced to 60 minutes. In the first step the mAb were cleaved using 1 unit of FabRICATOR ® /μg IgG (0.5-10 μg IgG / μl). After 30 min incubation at 37°C the reduction agents TCEP and Gd-HCl were added to a final concentration of 50 mM and 4 M, respectively and incubated for 30 min in room temperature. Samples were investigated by LC-MS using standard chromatography including desalting, combined with an UHR-ESI-QTOF (maXis 4G, Bruker) additionally automated acquisition, analysis and result generation, BioPharma Compass (Bruker) was used.
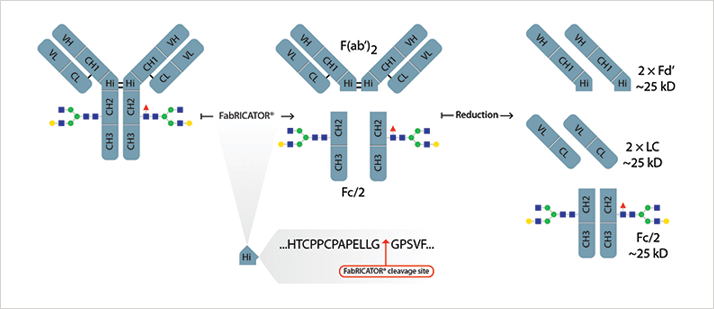
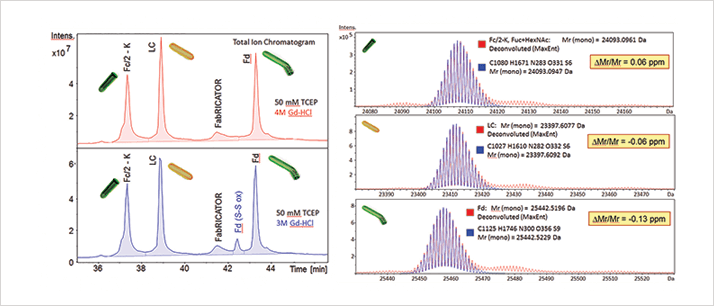
Using the presented protocol, complete workflows of sample preparation, data acquisition, data analysis, and report generation can be automated saving time and money. Additionally, compared to traditional peptide mapping the short incubation at neutral pH and limited number of steps reduces the risk of sample preparation induced modifications commonly observed with tryptic digests.
Please visit us at ASMS booth 126