A new spatial proteomics framework, PathoPlex, enables highly multiplexed protein imaging at subcellular resolution combined with computational pixel-based clustering. Developed by a team from the University of Aarhus and University of Hamburg (and collaborators), the approach addresses limitations in existing protein spatial analysis, particularly in complex tissues where multiple cell types interact in close proximity.
Using PathoPlex, the team analyzed kidney tissue samples, grouping pixels with similar protein signatures to map biological processes beyond the boundaries of individual cells. This method uncovered unexpected patterns, including JUN signalling activity in parietal epithelial cells and TRPC6 expression in tubular cells from patients with advanced diabetic kidney disease – findings that challenge existing assumptions about disease-related protein distribution.
We spoke with Malte Kuehl, co-first author of the study, to learn more about the technology, its applications, and future developments.

What was your main inspiration for this study?
In recent years, remarkable progress has been made in spatial transcriptomics, but on the protein side, solutions have remained limited. We fundamentally believe that examining proteins in their spatial context at high resolution – including extra- and sub-cellular analyses – is essential for understanding disease, and remains an underexplored area within multimodal experimental setups. This conviction stems partly from our background researching kidney tissues, where many different cell types interact in close spatial proximity – necessitating high-resolution imaging. It also stems from our experience with immune-mediated and fibrotic diseases, where both intracellular signalling and extracellular signals play a key role. These two aspects are not fully covered by the most common spatial proteomics analysis pipelines and is something we sought to improve with PathoPlex.
A lot of people are excited about spatial proteomics – it was named Method of the Year 2024 by Nature. But do you feel there are some limitations that need to be addressed?
Although spatial proteomics has advanced significantly, the focus has been partly on replicating insights from spatial transcriptomics, rather than exploring the unique potential of proteins. We believe there is a significant opportunity to progress from cellular-level to multi-scale analyses which complement RNA-based methods by quantifying structures, spatial organization, proteins and post-translational modifications that are invisible to them. Furthermore, the potential of multimodal approaches which combine not only RNA and proteins, but also histopathological morphology, metabolomics and other factors, remains largely unexplored. That said, in order to realize this potential, it will be necessary to establish quality control standards and ontologies for describing results, as well as interoperable software ecosystems – all of which are still in their infancy (though promising research is happening all around the world).
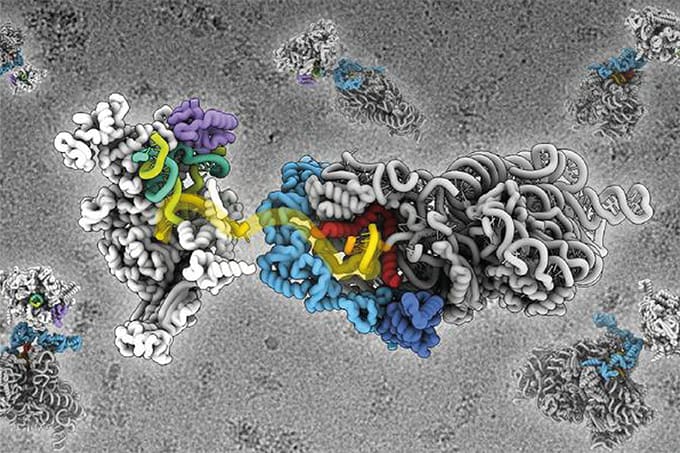
Could you briefly explain what PathoPlex is, and how it combines high-resolution imaging with pixel-based clustering to uncover biological insights from tissue?
PathoPlex is a framework that combines highly multiplexed protein imaging at subcellular resolution with a computational pipeline to analyze protein co-expression patterns. Rather than analyzing individual cells, our analysis focuses on pixel-based clustering to group pixels with similar protein signatures. This enables us to identify and spatially map biological processes with great resolution, providing a different, complementary perspective to protein biology.
What were the biggest technical or computational challenges you had to overcome – and how did you address them?
In the wet lab, we had to address issues such as tissue lifting, as well as simplifying liquid handling to enable scalability across dozens of imaging cycles, which we achieved using a specialized coating agent and 3D-printed plates. In terms of computational analyses, our terabyte-sized datasets exceeded the capacity of CPU-based solutions, so we developed our own Python library, “spatiomic,” to leverage GPU acceleration.
Did any of your findings surprise you? Any “eureka” moments?
The project was less about a single “eureka” moment, and more the process of seeing a complex puzzle slowly come together piece by piece. Although our initial antibody panel designs were based on existing knowledge to some degree, the spatial data often revealed unexpected findings. For instance, although JUN signalling had previously been linked to immune-mediated kidney disease, it was believed to be primarily active in immune cells. However, by combining our spatial resolution with cell-type markers, we discovered that it was actually a key switch in a different cell type: parietal epithelial cells. Similarly, we found that a protein implicated in podocytes in other kidney diseases, TRPC6, is actually most highly expressed in tubular cells in patients with advanced diabetic kidney disease. These discoveries were often the result of collaborative efforts, with experts from various institutions contributing their unique insights and materials to form a complete picture – as well as throwing up some surprises.
What are the most exciting potential applications of this framework?
PathoPlex is a foundational technology for the exploration of proteins within tissues and as such has a wide range of applications, but one particularly exciting one to me is in the study of rare diseases. It provides the necessary subcellular resolution and a sample-sparing workflow, potentially enabling researchers to obtain more data from very few biopsies and overcome barriers to studying neglected diseases.
Can you comment on the value of cross-disciplinary collaboration in advancing pathology and precision medicine – and where you see analytical science playing the most important role?
In the current scientific landscape, studies are often so complex that no individual research group can master all the necessary techniques, from maintaining biobanks and performing advanced microscopy to developing high-quality software. Collaborating with a large, international and interdisciplinary team was the best way we could address a problem of this scale and it was an honour to be part of such a great team with diverse expertise. Analytical science provided the framework for our study, enabling us to generate and process multilayered datasets. This ability to generate, integrate and analyze complex data to gain insights into disease is ultimately the frontier on the path towards precision medicine.
What are your next steps for expanding the PathoPlex framework – either in terms of new disease areas, automation, or integration with other omics technologies?
We are collaborating with multiple partners to apply PathoPlex to new disease areas and organs, as we believe it can provide valuable insights into a broad range of pathologies. A key focus is the integration of additional data modalities from both laboratory and clinical settings. Technically, we are working to connect different scales of analysis including deep learning-based segmentation and pixel-level clustering. We are also seeking to get more out of the spatial nature of our data through additional spatial statistics applications. Lastly, to assist with the planning and interpretation of complex experiments – tasks which can require weeks of research – we recently launched biocontext.ai: a community initiative to connect biomedical tools with agentic AI systems to help researchers access relevant biomedical knowledge, connect data across sources and – hopefully soon – use these new technologies to better scale their work.
Malte Kuehl is a Postdoctoral Researcher in the Department of Clinical Medicine, Section of Pathology, at Aarhus University